I would like to encourage all of our male Wilson cousins to seriously consider taking or upgrading to the Family Tree DNA Big-Y test. As it is, I am the only one in our Y-line (direct male) going back over 2000 years to have taken this particular test — it’s lonely out there! Note that, as I have explained in another post, this is a different test than the Y-xx tests that a number of male Wilsons have taken. The “standard” Y tests measure STR’s (Short Tandem Repeats) which vary much more quickly and thus give more refined matches in genealogical times, whereas the Big-Y looks at SNP’s (Single Nucleotide Polymorphisms) which only mutate, on average, about once every 80-150 years. Thus, across all of James Sr’s descendants over almost 300 years, there might be only 2-3 mutations in any given line (and of course, they would all be different). However, as this level of DNA testing is still in its infancy, the frequency of SNP mutations is being debated and developed – previous estimates of 144 years/per mutation (on average) are now being reconsidered with values such as 84 and even 44 being proposed. This actually presents an opportunity for our own science experiment, which I’ll describe more later.
As I write this (June 14, 2020) Family Tree DNA is having their annual Father’s Day sale on Y-DNA tests. It is admittedly difficult to give a compelling rationale as to why anyone should spend their hard-earned bucks on more DNA testing of this type. After all, it is likely that many of us have already had at least 1 autosomal test done (such as at Ancestry or 23andme) , and several of us have also done one or more Y-xxx tests. And, in some of our cases, we know exactly how we are related — for instance, I know of a male 3rd cousin that is also related to James C. Wilson, my g-g-grandfather. If he were to upgrade to the Big-Y test, the results wouldn’t give us any more information about how were related since we already have the paper trail. For the cousins whose distant parentage is less established (such as if they were descended from one of the many Moses, or from John Culpepper or Daniel Wilson, neither of whom we know exactly how was related to James Sr.), the difference of only 2-3 SNPs that would be likely in this time-frame wouldn’t provide enough resolution to identify a specific relationship or generation.
However, we could use these results to start building our own genetic Wilson family tree and our distinct Wilson branch. Let’s assume that James Sr’s haplogroup was R-J100 (my made-up name) which would have been defined by his particular set of mutations (which he may or may not have shared with his father or any brothers or cousins). Any of his sons might have the exact same haplogroup (i.e. SNPs), or one of more of them might have their own mutations. Eventually along each of their lines, a few mutations will occur and will all be different depending on which son/grandson/greatgrandson/etc. was the patriarch of his line. However, every single one of us would have J100, plus maybe a few of our own. So, R-J100 would define the root of our own branch. That would be establish (by FTDNA) by identifying the last common SNP that all of us share, assuming enough of our branches took the Big-Y test. Then, we could group the SNP’s that belong, for instance, to Thomas Sr’s line, as well as those of John Culpepper and Daniel, and others. Even with only 2 or 3 mutations we might be able to start seeing signatures for each line, which would then help us place “newcomers” to the testing results closer to the correct lineage. See below diagram which I hope illustrates this.
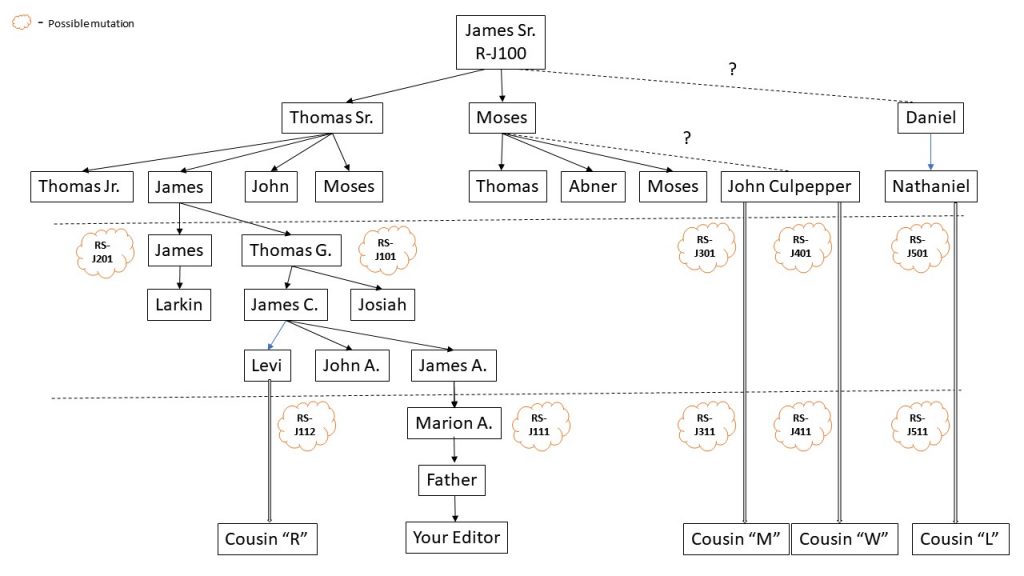
Even more importantly, I think, would be our ability to better identify James Sr.’s own ancestors. For instance, if a male that was descended from one of James Scottish (I presume) ancestors were tested, we would see a branch that occurred before James’ time, and be able to to roughly identify a time-frame for the common ancestor (assuming enough of us had taken the Big-Y to establish the branching point for James Sr.). As the testing result database grows, we would perhaps start to see more and more of these “cousins”, and if and of them had good paper genealogy trails we might get a much firmer grasp on James’ ancestry and familial origins.
Finally, the really fun thing would be to establish our own dataset for the research of mutation frequency. Given that most of us already know how many generations we are removed from James Sr., give or take a generation, developing a sufficiently large set of variant values would provide an average mutation frequency we could use to add to the research. For example, once the haplogroup for James Sr. was established, if most of us had, say, around 6 variants from that, the mutation frequency would be ~42 years (250 years/6). That would be a major finding that differs greatly from the 80-150 years currently being assumed. Or, we might find we really do have 2-3 on average, which would substantiate the existing estimates. Either way, we could provide our own Wilson group contribution to genetic science. I think that would be pretty cool!
